This web page was produced as an assignment for Gen677 at UW-Madison Spring 2010
Future Directions
There is much about CML that is not understood. Areas of research include but not limited to the mechanism of the translocation of the Philadelphia, regulation of BCR-ABL, the molecular pathway of BCR-ABL, and possible treatments using drugs to target the BCR-ABL protein and stem cells containing the Philadelphia chromosome.
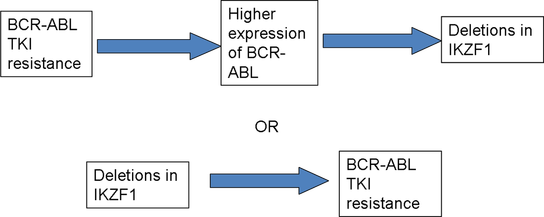
One of the more recent discoveries is the addition of deletion or insertion of other genes not directly associated with BCR-ABL. An example of this is found in the deletion of the IKZF1 gene, which transcribes a transcription factor that regulates cell differentiation of B & T cells. These gene alterations have been seen in patients that have acquired resistance to TKIs and in the phase transition from the chronic phase to the blast crisis stage. What is not known is what occurs first: the TKI resistance or the gene alternations that lead to the phase transition.
One of the more recent discoveries is the addition of deletion or insertion of other genes not directly associated with BCR-ABL. An example of this is found in the deletion of the IKZF1 gene, which transcribes a transcription factor that regulates cell differentiation of B & T cells. These gene alterations have been seen in patients that have acquired resistance to TKIs and in the phase transition from the chronic phase to the blast crisis stage. What is not known is what occurs first: the TKI resistance or the gene alternations that lead to the phase transition.
Proposed Experiment
Question: What is the molecular pathway of BCR-ABL and IKZF1?
Hypothesis: TKI resistance allows the overexpression of BCR-ABL, increasing genomic instability and resulting in the deletion in the IKZF1 gene.
Experiment:
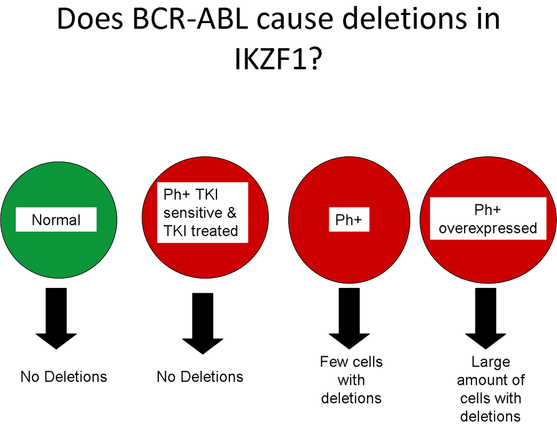
Phase 1- Does BCR-ABL cause deletions in IKZF1?
In order to determine whether BCR-ABL causes deletions in the IKZF1 gene, I would take four groups of human culture myeloid stem cells. The first group, the contol group, would be normal myeloid stem cells from a healthy individual negative for CML. The next three groups are the experimental groups all containing the Philadelphia chromosome and therefore contain the BCR-ABL fusion gene. The first experimental group is sensitive to TKIs and will be treated with TKIs. The second group will not have any alterations to the cell and the third group will have the the BCR-ABL fusion gene overexpressed.
After a pre-determined time period, a CGH array will be performed to detect deletions in the IKZF1 gene.
If my hypothesis is correct, the control group and the TKI treated experimental group will contain no deletions in the IKZF1 gene. The other two experimental groups will contain deletions, but the overexpressed BCR-ABL experimental group will have more cells with the deletion than the experimental groups. This provides evidence that the expression of BCR-ABL seen in TKI resistance leads to the deletion of the IKZF1 gene.
Experiment:
Phase 1- Does BCR-ABL cause deletions in IKZF1?
In order to determine whether BCR-ABL causes deletions in the IKZF1 gene, I would take four groups of human culture myeloid stem cells. The first group, the contol group, would be normal myeloid stem cells from a healthy individual negative for CML. The next three groups are the experimental groups all containing the Philadelphia chromosome and therefore contain the BCR-ABL fusion gene. The first experimental group is sensitive to TKIs and will be treated with TKIs. The second group will not have any alterations to the cell and the third group will have the the BCR-ABL fusion gene overexpressed.
After a pre-determined time period, a CGH array will be performed to detect deletions in the IKZF1 gene.
If my hypothesis is correct, the control group and the TKI treated experimental group will contain no deletions in the IKZF1 gene. The other two experimental groups will contain deletions, but the overexpressed BCR-ABL experimental group will have more cells with the deletion than the experimental groups. This provides evidence that the expression of BCR-ABL seen in TKI resistance leads to the deletion of the IKZF1 gene.
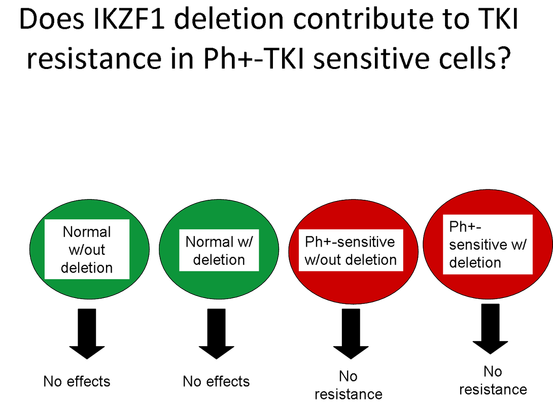
Phase 2 - Does IKZF1 deletion contribute to TKI resistance in Ph+-TKI sensitive cells?
The second phase of the experiment provids evidence that the other theory is incorrect and the deletion in the IKZF1 gene does not cause TKI resistance. Like the first phase of the experiment, human cultured myeloid stem cells will used. There will be two control groups and two experimental groups. The two control groups will consist of normal myeloid stem cells from health individuals and one of those groups will have the IKZF1 gene removed. The two experimental will have the Philadelphia chromosome containing the BCR-ABL fusion gene which is sensitive to TKIs. One of these two groups will have the deletion of IKZF1 gene. After a predetermined time period, the cell cultures will be treated with TKIs and examined for resistance through cell proliferation rate and the amount of BCR-ABL protein produced using the western blot.
If my hypothesis is correct, TKI resistance will not form in any of the experimental groups
Phase 3 - What genes are overexpressed in Ph+ cells with IKZF1 deletion?
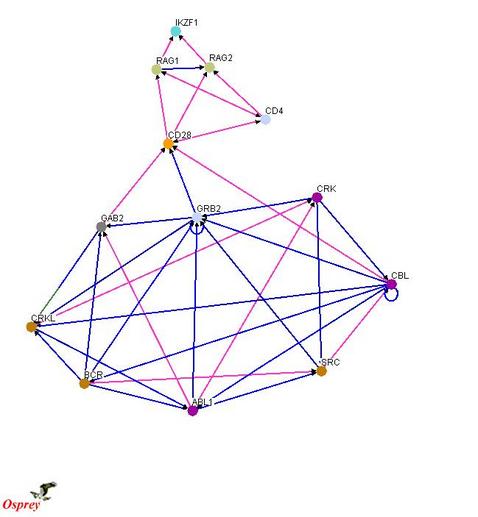
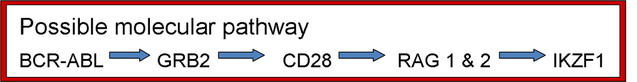
The purpose of this part of the experiment is to determine what genes and their protein products are involved in the formation of the deletion in the IKZF1 gene. There has been evidence that the deletion is a result of the stimulation of the RAG complex. In order to form a hypothesis of what genes and proteins may be involved, I looked at the protein-protein interactions for ABL1, IKZF1, and RAG and find a connection between the interaction networks. Below is a possible protein-protein interactions network that connects ABL1 and IKZF1 through the RAG complex.
A microarray comparing the expression level of TKI-treated human myeloid stem cells having the BCR-ABL fusion gene and its TKI sensitive protein product to human myeloid stem cells having the BCR-ABL fusion gene and the acquired IKZF1 deletion. The expected results would be increased expression of GRB2, CD28, and proteins involved in the RAG complex. There may also be increased expression of other proteins seen in the theortical protein-protein interaction network pictured above. This provides insight into what the molecular pathway may be that causes the deletions in the IKZF1 gene.
Further Research
Further research based on the results of this experiment include:
- investigate into the molecular pathway by controlling the expression of genes believed to be involved in the pathway in normal cells and determining if the deletion in the IKZF1 still occurs and by exploring protein complexes that involve the upregulated genes
- determine possible treatments to prevent deletion and decrease risk of disease progression
- investigate other gene alterations
References
1. STRING http://string.embl.de/
2. Osprey http://biodata.mshri.on.ca/osprey/servlet/Index
3. Joha, S., Dauphin, V., Fepretre, F. et al. (2011). Genomic characterization of imatinib resistance in CD34+ cell populations from chronic myeloid eukemia patients. Leukemia Research, 35, 448-458.
4. Nacheva, E., Brazma, D., Virgili, A. et al. (2010). Deletions of immunoglobulin heavy chain and T cell receptor gene regions are uniquely associated with lymphoid blast transformation of chronic myeloid leukemia
2. Osprey http://biodata.mshri.on.ca/osprey/servlet/Index
3. Joha, S., Dauphin, V., Fepretre, F. et al. (2011). Genomic characterization of imatinib resistance in CD34+ cell populations from chronic myeloid eukemia patients. Leukemia Research, 35, 448-458.
4. Nacheva, E., Brazma, D., Virgili, A. et al. (2010). Deletions of immunoglobulin heavy chain and T cell receptor gene regions are uniquely associated with lymphoid blast transformation of chronic myeloid leukemia