This web page was produced as an assignment for Gen677 at UW-Madison Spring 2010
Microarray
Microarray data involving CML and the human ABL1 gene was obtained using the NCBI GEO. Microarrays are useful in comparing gene expression under two different conditions including examples such as a normal gene vs. a mutated gene or a untreated cell line vs. a treated cell line.
Chronic phase chronic myelogenous leukemia: CD34+ hematopoietic stem and progenitor cells
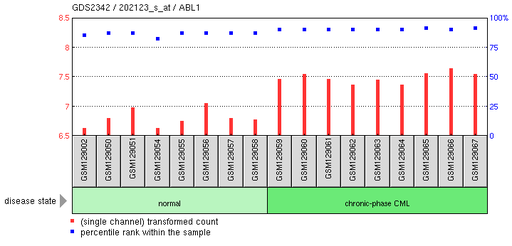
This experiment compared the gene expression of normal CD34+ cells with CML CD34+ cells isolated in the chronic stage. There was an increase in expression of the ABL1 gene in the chronic-phase CML patients compared to healthy individuals.
Imatinib effects on chronic myelogenous leukemia CD34+ cells
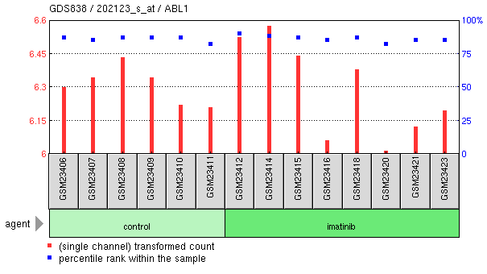
This experiment compared the expression level of ABL1 in healthy donors and imatinib-treated CML patients. The results of the study showed there to be no difference in gene expression of ABL1 between the two conditions. It was originally thought that imatinib inhibited BCR-ABL and wild-type ABL1.
Cyclophosphamide-resistant chronic myelogenous leukemia cell line
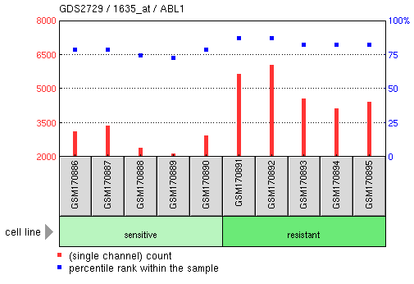
This experiment compared gene expression of cyclophosphamide-resistant leukemia cell lines to cyclophosphamide-sensitive cell lines. Cyclophosphamide is another possible treatment for CML. The microarray showed an increase in the the ABL1 gene expression in the resistant cell lines.
Analysis
Microarrays have been useful in studying CML even though the focus of majority of these studies have not placed emphasis on the expression of ABL1 gene. These experiments have used microarrays to determine the molecular pathways of CML and current and possible treatments in hope to understand how the disease evolves and new targets to prevent the CML from progressing to the later and more fatal stages.
